Slowing DNA Transport Using Graphene–DNA Interactions
A molecular dynamics simulation modeling the atomistic details of single stranded DNA (ssDNA) translocating through a nanopore emended in a stacked graphene-dielectric-graphene membrane. The individual nucleotides (nt) of the 54 nt ssDNA molecule are uniquely colored while the membrane is translucent for clarity. The movie illustrates a cross-sectional (left) and top-down (right) view of the ssDNA translocation process driven by a 500 mV transmembrane potential.
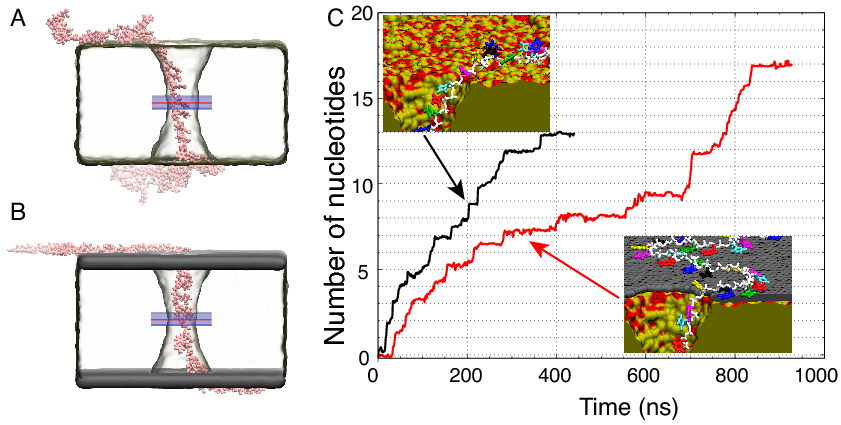
An insight into the mechanics of the single-stranded DNA translocation through a dielectric (A) and graphene-dielectric-graphene stacked nanopore (B). Hydrophobic stacking of the ssDNA bases onto the graphene sheets layered above and below the dielectric reduces the translocation rate of the ssDNA through the stacked membrane (C, red) compared to just a dielectric membrane (C, black).
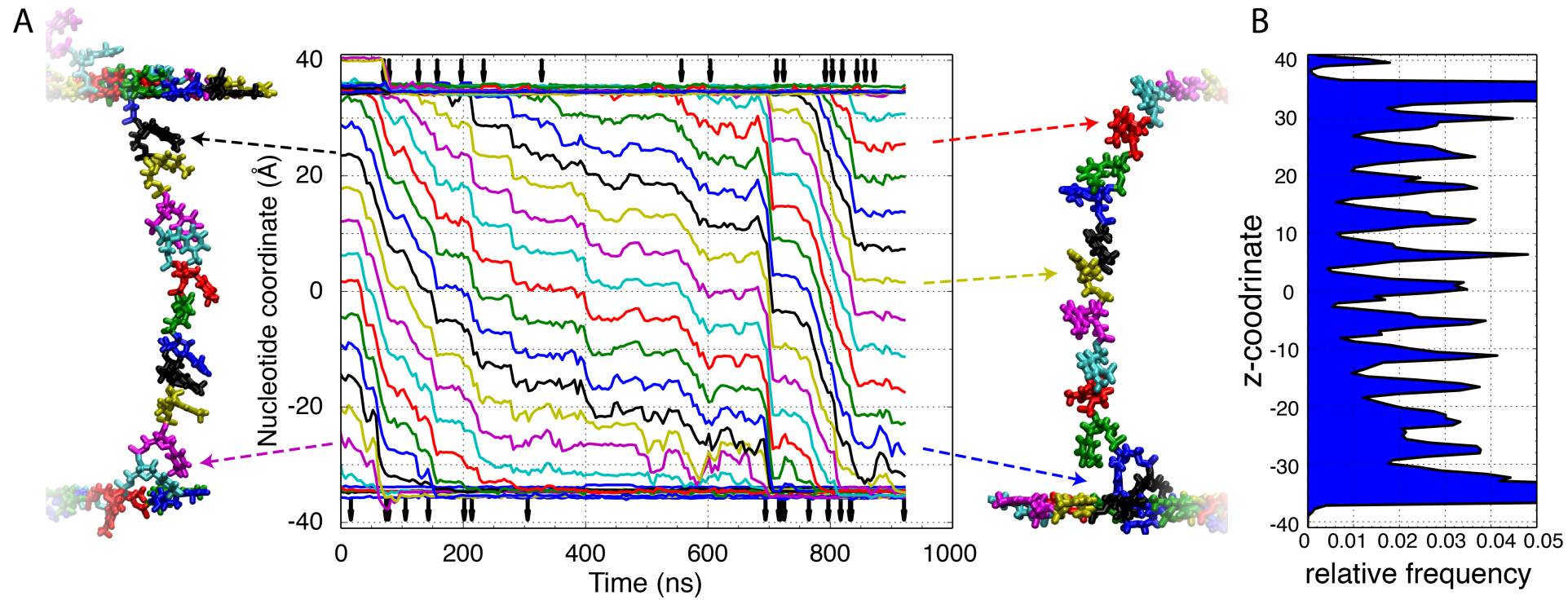
The ssDNA translocates through the stacked membrane in a stepwise manner. The z-coordinate of each individual nucleotide (nt) measured as the center of mass of each nucleotides backbone (A). Each nucleotide's color corresponds to the same color trace. Arrows above and below the traces indicate the moments of DNA base unbinding from the top layer of graphene (top) and binds to the bottom layer of graphene (bottom). The traces illustrate periods of very little motion punctuated by short quick movements approximately the length of one nucleotide. A distribution of the locations of the ssDNA nucleotide along the z-coordinate indicate repetitive placement of the ssDNA nucleotides within the same region of the pore.